Last week's molecule was the drug pantoprazole, a proton pump inhibitor used to treat excess stomach acid or acid reflux [Monday's Molecule #225 ]. The winner is Bill Gunn.
This week's molecule (left) is related to one from last April. That molecule, is one of the essential molecules in the human diet and today's molecule is the reason why. This is one of those molecules that everyone should recognize because it's a key metabolic precursor in a large number of species. This is one of those times when all you have to do is supply the common name (Merry Christmas!) and NOT the IUPAC systematic name that correctly identifies the exact molecule shown in the image. However, if anyone wants to supply the systemiac name, feel free to do so.
Email your answer to me at: Monday's Molecule #227. I'll hold off posting your answers for at least 24 hours. The first one with the correct answer wins. I will only post the names of people with mostly correct answers to avoid embarrassment. The winner will be treated to a free lunch.
There could be two winners. If the first correct answer isn't from an undergraduate student then I'll select a second winner from those undergraduates who post the correct answer. You will need to identify yourself as an undergraduate in order to win. (Put "undergraduate" at the bottom of your email message.)
Read more »
Showing posts with label Biochemistry. Show all posts
Showing posts with label Biochemistry. Show all posts
Monday, December 16, 2013
Monday, December 9, 2013
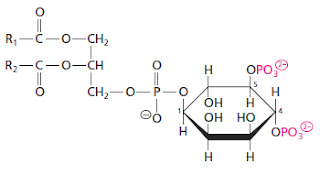
Monday's Molecule #226
Last week's molecule was sucrose 6-phosphate or α-D-glucopyranosyl-(1→2)-β-D-fructofuranoside 6-phosphate [Monday's Molecule #225 ]. The winner is Jean-Marc Neuhaus (again). He appears to be the only Sandwalk reader who has a copy of my book!
Today's molecule (below) looks a bit strange. It should be obvious that this is not a "natural" molecule. What is it and what does it do? You don't need to give me a long systematic name. The common name will do quite nicely.
Email your answer to me at: Monday's Molecule #226. I'll hold off posting your answers for at least 24 hours. The first one with the correct answer wins. I will only post the names of people with mostly correct answers to avoid embarrassment. The winner will be treated to a free lunch.
There could be two winners. If the first correct answer isn't from an undergraduate student then I'll select a second winner from those undergraduates who post the correct answer. You will need to identify yourself as an undergraduate in order to win. (Put "undergraduate" at the bottom of your email message.)
Read more »
Today's molecule (below) looks a bit strange. It should be obvious that this is not a "natural" molecule. What is it and what does it do? You don't need to give me a long systematic name. The common name will do quite nicely.
Email your answer to me at: Monday's Molecule #226. I'll hold off posting your answers for at least 24 hours. The first one with the correct answer wins. I will only post the names of people with mostly correct answers to avoid embarrassment. The winner will be treated to a free lunch.
There could be two winners. If the first correct answer isn't from an undergraduate student then I'll select a second winner from those undergraduates who post the correct answer. You will need to identify yourself as an undergraduate in order to win. (Put "undergraduate" at the bottom of your email message.)
Read more »
Tuesday, December 3, 2013
Lenski's long-term evolution experiment: the evolution of bacteria that can use citrate as a carbon source
Richard Lenski set up twelve flasks of E. coli B back in 1988. They were allowed to grow overnight in minimal medium containing 139μM glucose and 1,700 μM citrate. The glucose was the only carbon source that the parent strain could use and it limited growth of the cultures. The citrate was present as a standard chelating agent. The bacteria could not take up citrate and use it as an additional carbon source.
Every day the culture was diluted by transferring one ml to 99 ml of fresh medium (1/100). There were 6.64 generations per day or 2,423 generations per year (slightly more in a leap year).
These cultures were under strong selective pressure. Individual bacteria that could grow faster would out-compete other bacteria in the culture and take over. Lenski expected that each culture would show a variety of different solutions to the selective pressure with many common mutations and many that could be unusual.
Read more »
Every day the culture was diluted by transferring one ml to 99 ml of fresh medium (1/100). There were 6.64 generations per day or 2,423 generations per year (slightly more in a leap year).
These cultures were under strong selective pressure. Individual bacteria that could grow faster would out-compete other bacteria in the culture and take over. Lenski expected that each culture would show a variety of different solutions to the selective pressure with many common mutations and many that could be unusual.
Read more »
Monday, December 2, 2013
Monday's Molecule #225
Last week's molecule was EF-Tu (elongation factor-thermo unstable). EF-Tu binds to all tRNA molecules in the cell and helps position them in the A-site of the ribosome-mRNA complex during protein synthesis. Its release is coupled to GTP hydrolysis. Eukaryotes contain homologous proteins with different names (e.g. EF-1α). The winner was Jon Nuelle from Texas. The undergraduate winner was Ariel Gershon for the second week in a row.
Today's molecule (below) is an intermediate in a very important pathway that's only found in some species. This is one of those times when you need to supply the common name AND the correct scientific name that specifies the exact molecule shown in the figure.
Email your answer to me at: Monday's Molecule #225. I'll hold off posting your answers for at least 24 hours. The first one with the correct answer wins. I will only post the names of people with mostly correct answers to avoid embarrassment. The winner will be treated to a free lunch.
There could be two winners. If the first correct answer isn't from an undergraduate student then I'll select a second winner from those undergraduates who post the correct answer. You will need to identify yourself as an undergraduate in order to win. (Put "undergraduate" at the bottom of your email message.)
Read more »
Today's molecule (below) is an intermediate in a very important pathway that's only found in some species. This is one of those times when you need to supply the common name AND the correct scientific name that specifies the exact molecule shown in the figure.
Email your answer to me at: Monday's Molecule #225. I'll hold off posting your answers for at least 24 hours. The first one with the correct answer wins. I will only post the names of people with mostly correct answers to avoid embarrassment. The winner will be treated to a free lunch.
There could be two winners. If the first correct answer isn't from an undergraduate student then I'll select a second winner from those undergraduates who post the correct answer. You will need to identify yourself as an undergraduate in order to win. (Put "undergraduate" at the bottom of your email message.)
Read more »
Sunday, December 1, 2013
Elizabeth Pennisi writes about Richard Lenski's long-term evolution experiment
Elizabeth Pennisi has written about the long-term evolution experiment of Richard Lenski [The Man Who Bottled Evolution]. The experiment is in it's 25th year and it is entirely appropriate that Science magazine devotes several pages to describing the results. There have been some remarkable discoveries.
But I want to focus on a couple of things that Pennisi says in her article. There has also been a discussion on Panda's Thunb: Lenski’s experiment: 25 years and 58,000 generations. Pennisi writes ...
Read more »
But I want to focus on a couple of things that Pennisi says in her article. There has also been a discussion on Panda's Thunb: Lenski’s experiment: 25 years and 58,000 generations. Pennisi writes ...
Lenski's humble E. coli have shown, among other things, how multiple small mutations can prepare the ground for a major change; how new species can arise and diverge; and that Gould was mistaken when he claimed that, given a second chance, evolution would likely take a completely different course. Most recently, the colonies have demonstrated that, contrary to what many biologists thought, evolution never comes to a stop, even in an unchanging environment.Let's talk about two issues in that paragraph.
Read more »
Monday, November 25, 2013
Monday's Molecule #224
Last week's molecule was the second messenger, phosphatidylinositol 4,5-bisphosphate or PIP2. The winner is Dean Bruce. The undergraduate winner is Ariel Gershon [Monday's Molecule #223].
Today's molecule is a protein (purple). It's one of the most abundant proteins in E. coli because it's bound to almost all tRNA molecules in the cell. Name the protein (complete name, not just initials).
Email your answer to me at: Monday's Molecule #224. I'll hold off posting your answers for at least 24 hours. The first one with the correct answer wins. I will only post the names of people with mostly correct answers to avoid embarrassment. The winner will be treated to a free lunch.
There could be two winners. If the first correct answer isn't from an undergraduate student then I'll select a second winner from those undergraduates who post the correct answer. You will need to identify yourself as an undergraduate in order to win. (Put "undergraduate" at the bottom of your email message.)
Read more »
Today's molecule is a protein (purple). It's one of the most abundant proteins in E. coli because it's bound to almost all tRNA molecules in the cell. Name the protein (complete name, not just initials).
Email your answer to me at: Monday's Molecule #224. I'll hold off posting your answers for at least 24 hours. The first one with the correct answer wins. I will only post the names of people with mostly correct answers to avoid embarrassment. The winner will be treated to a free lunch.
There could be two winners. If the first correct answer isn't from an undergraduate student then I'll select a second winner from those undergraduates who post the correct answer. You will need to identify yourself as an undergraduate in order to win. (Put "undergraduate" at the bottom of your email message.)
Read more »
Wednesday, November 20, 2013
Fred Sanger (1918-2013)
BBC News is reporting that Fred Sangerhas died [Frederick Sanger: Double Nobel Prize winner dies at 95]. Sanger is one of the few people to win two Nobel Prizes. His first was for sequencing insulin and his second was for developing a technique for sequencing DNA (Sanger sequencing).
Most people, even most scientists, have no idea how much he influenced molecular biology. Sanger worked at Cambridge (UK). When Francis Crick first arrived at Cambridge in 1947 he soon met a number of important scientists. Here's how Horace Freeland Judson describes Sanger in The Eight Day of Creation (pp. 88-89).
Most people, even most scientists, have no idea how much he influenced molecular biology. Sanger worked at Cambridge (UK). When Francis Crick first arrived at Cambridge in 1947 he soon met a number of important scientists. Here's how Horace Freeland Judson describes Sanger in The Eight Day of Creation (pp. 88-89).
One of these in particular, the biochemist Frederick Sanger, came to have great intellectual importance in Crick's thinking and then to molecular biologists generally as the field developed. Sanger is temperamentally and in scientific style Crick's opposite. Where many scientists, Crick among them, flower at conferences and do a great deal of their science by talking, Singer is a quiet man—reticent, even shy, a man who worked with his hands, at the bench. He almost never talked to the press, never despite the editor's importuning wrote the big article for Scientific American. One might spot him bicycling to work on a spring morning, in a drab brown coat, in the rain. Once I stopped to talk with him in the corridor of the laboratory building, where he was waiting in the queue for his turn at the ultraviolet-light box, in order to illuminate the spots on a sheet of chromatography paper he was holding. Sanger is a Quaker by upbringing, and stayed at Cambridge through the second world war; holding only a junior fellowship in the biochemistry department, and even when the war dried up the usual sources of research funds, with family money he was able to keep going. In the course of nearly a decade, beginning in the mid-forties, Sanger settled upon the new techniques of chromatography to determine the amino-acid sequences of the two chains of the bovine insulin molecule. He proved that the sequences are unique and always the same, meaning that every molecule of insulin in every cow is exactly like every other. Yet the sequences show no general periodicities: they are not predictable from ordinary chemical rules.
Sanger published very rarely. His papers came to be red with heart in mouth by other scientists, for they are technically brilliant. Even as he worked, though, the news slowly spread and the implications sank in. For one thing, his department held a biochemistry tea club where perhaps once a month research that was relatively finished, though not yet submitted for publication, was presented. Brigitte Askonas, later an important figure in immunology in England, came to Sanger's lab as a doctoral student late in 1948, staying on into 1952. "Even then, Fred had only a minor fellowship—and some had wanted to kick him out," she told me once. "When one would ask him how his work was going, he would say very little. 'Oh, I've got another peptide.'" Then at a lab meeting he would bring a stack of cards showing overlapping short sequences, and slowly, diffidently, build up his latest segment of the molecule. "Crick always came to the tea club," Askonas said. "And he always asked awkward questions. Enfant terrible questions. And then he would explain, somewhat disingenuously, 'You see, I'm just learning.'" Sanger's general conclusion was forceful by 1949, when he went to the annual symposium on quantitative biology at Cold Spring Harbor (his only such visit). In a paper published on the first of June of that year—the earliest of his magisterial series of papers on insulin appearing every odd-numbered year until 1955—he was already able to say that "there appears to be no principle that defines the nature of the [amino-acid] residue" occupying any particular position in a protein. The conclusion was definitive by 1951. For this work and the methods of sequencing he invented to do it, Sanger was awarded the Nobel Prize in chemistry in 1958. (He later turned to the more difficult problem of sequencing nucleic acids, which earned him a share of another Nobel Prize, in 1980. Crick, from his first arrival in Cambridge, new of Sanger's work step by step, months and even years before new steps were published.
Monday, November 18, 2013
Monday's Molecule #223
Last week's molecule was a Holliday junction, one of the key intermediates in recombination. It's named after Robin Holliday who has since retired from science to concentrate on being a sculptor. He has produced several "biological" sculptures including "DNA Structure" (top) and "Cross Over" (bottom). The winner is Caroline Josefsson from British Columbia. The undergraduate winner is Andrew Wallace but since he lives in Australia, I suspect he won't be coming to lunch [Monday's Molecule #222].
Today's molecule (below) is not one of my favorite molecules for many reasons. However, it's pretty important in some species. Name the molecule, being as specific as you can without resorting to IUPAC rules. I need the most common name as well as a more detailed name.
Email your answer to me at: Monday's Molecule #223. I'll hold off posting your answers for at least 24 hours. The first one with the correct answer wins. I will only post the names of people with mostly correct answers to avoid embarrassment. The winner will be treated to a free lunch.
There could be two winners. If the first correct answer isn't from an undergraduate student then I'll select a second winner from those undergraduates who post the correct answer. You will need to identify yourself as an undergraduate in order to win. (Put "undergraduate" at the bottom of your email message.)
Read more »
Today's molecule (below) is not one of my favorite molecules for many reasons. However, it's pretty important in some species. Name the molecule, being as specific as you can without resorting to IUPAC rules. I need the most common name as well as a more detailed name.
Email your answer to me at: Monday's Molecule #223. I'll hold off posting your answers for at least 24 hours. The first one with the correct answer wins. I will only post the names of people with mostly correct answers to avoid embarrassment. The winner will be treated to a free lunch.
There could be two winners. If the first correct answer isn't from an undergraduate student then I'll select a second winner from those undergraduates who post the correct answer. You will need to identify yourself as an undergraduate in order to win. (Put "undergraduate" at the bottom of your email message.)
Read more »
Monday, November 11, 2013
Monday's Molecule #222
Last week's molecule was D-serine. (Not L-serine.) The winner is undergraduate Zhimeng Yu [Monday's Molecule #221].
I was reminded of this week's molecule by a discussion we are having in an evolution forum and by a comment from a student who took a MOOC on genetics. Does it depict something that should be taught in every introductory genetics course? Is it something that should be discussed in an evolution course? You need to name the structure formed by the blue, gray, and black strands. It has a specific name.
Email your answer to me at: Monday's Molecule #222. I'll hold off posting your answers for at least 24 hours. The first one with the correct answer wins. I will only post the names of people with mostly correct answers to avoid embarrassment. The winner will be treated to a free lunch.
There could be two winners. If the first correct answer isn't from an undergraduate student then I'll select a second winner from those undergraduates who post the correct answer. You will need to identify yourself as an undergraduate in order to win. (Put "undergraduate" at the bottom of your email message.)
Read more »
I was reminded of this week's molecule by a discussion we are having in an evolution forum and by a comment from a student who took a MOOC on genetics. Does it depict something that should be taught in every introductory genetics course? Is it something that should be discussed in an evolution course? You need to name the structure formed by the blue, gray, and black strands. It has a specific name.
Email your answer to me at: Monday's Molecule #222. I'll hold off posting your answers for at least 24 hours. The first one with the correct answer wins. I will only post the names of people with mostly correct answers to avoid embarrassment. The winner will be treated to a free lunch.
There could be two winners. If the first correct answer isn't from an undergraduate student then I'll select a second winner from those undergraduates who post the correct answer. You will need to identify yourself as an undergraduate in order to win. (Put "undergraduate" at the bottom of your email message.)
Read more »
Tuesday, November 5, 2013
Test Your Scientific Skepticism
I once posted a series of articles on Roundup® (glyphosate) explaining how it works and how one makes Roundup®-Ready genetically modified plants.
Read more »
- How Roundup® Works
- Roundup Ready® Transgenic Plants
- The Molecular Basis of Roundup® Resistance
- Glyphosate-resistant Weeds
- Roundup® Is Safe
Read more »
Friday, November 1, 2013
Vertebrate Complexity Is Explained by the Evolution of Long-Range Interactions that Regulate Transcription?
The Deflated Ego Problem is a very serious problem in molecular biology. It refers to the fact that many molecular biologists were puzzled and upset to learn that humans have about the same number of genes as all other multicellular eukaryotes. The "problem" is often introduced by stating that the experts working on the human genome project expected at least 100,000 genes but were "shocked' when the first draft of the human genome showed only 30,000 genes (now down to about 25,000). This story is a myth as I document in: Facts and Myths Concerning the Historical Estimates of the Number of Genes in the Human Genome. Truth is, most knowledgeable experts expected that humans would have about the same number of genes as other animals. They realized that the differences between fruit flies and humans, for example, didn't depend on a host of new human genes but on the timing and expression of a mostly common set of genes.
This isn't good enough for many human chauvinists. They are still looking for something special that sets human apart from all other animals. I listed seven possibilities in my post on the deflated ego problem:
Read more »
This isn't good enough for many human chauvinists. They are still looking for something special that sets human apart from all other animals. I listed seven possibilities in my post on the deflated ego problem:
Read more »
Wednesday, October 30, 2013
Time to Re-Write the Textbooks! Nature Publishes a New Version of the Citric Acid Cycle
I was looking through my copy of Nature the other day trying to take seriously all the special reviews on "Transcription and Epigenetics." One article caught my eye ...
Gut, P. and Verdin, E. (2013) The nexus of chromatin regulation and intermediary metabolism. Nature 502:489-498. [doi: 10.1038/nature12752]
It's the figure showing the citric acid cycle (TCA cycle) that shocked me.
Textbooks show that the products of the citric acid cycle are ...
That's three NADH, one QH2, and one GTP (or ATP) for a total of ten ATP equivalents. The new version, published last week in the most prestigious science journal in the world, shows that there are six NADH produced per cycle for a total of 15 ATP equivalents. It must be correct because this is a paper about intermediary metabolism and it was reviewed by experts in the field. Unfortunately, the authors don't give a reference to this new information. I assume that it's common knowledge among the top metabolism researchers so they didn't bother citing the papers.
Can anyone out there direct me to the revolutionary papers that I missed?
P.S. I'm not even going to mention that FADH2 is NOT a product of enzyme-catalyzed β-oxidation.
Gut, P. and Verdin, E. (2013) The nexus of chromatin regulation and intermediary metabolism. Nature 502:489-498. [doi: 10.1038/nature12752]
Living organisms and individual cells continuously adapt to changes in their environment. Those changes are particularly sensitive to fluctuations in the availability of energy substrates. The cellular transcriptional machinery and its chromatin-associated proteins integrate environmental inputs to mediate homeostatic responses through gene regulation. Numerous connections between products of intermediary metabolism and chromatin proteins have recently been identified. Chromatin modifications that occur in response to metabolic signals are dynamic or stable and might even be inherited transgenerationally. These emerging concepts have biological relevance to tissue homeostasis, disease and ageing.The authors argue that, among other things, methylation of histones is regulated by changes in the concentrations of some citric acid cycle metabolites. I find it difficult to imagine that the concentrations of the citric acid cycle intermediates could change significantly enough to act as allosteric effectors but that's not what grabbed my attention.
It's the figure showing the citric acid cycle (TCA cycle) that shocked me.
Textbooks show that the products of the citric acid cycle are ...
That's three NADH, one QH2, and one GTP (or ATP) for a total of ten ATP equivalents. The new version, published last week in the most prestigious science journal in the world, shows that there are six NADH produced per cycle for a total of 15 ATP equivalents. It must be correct because this is a paper about intermediary metabolism and it was reviewed by experts in the field. Unfortunately, the authors don't give a reference to this new information. I assume that it's common knowledge among the top metabolism researchers so they didn't bother citing the papers.
Can anyone out there direct me to the revolutionary papers that I missed?
P.S. I'm not even going to mention that FADH2 is NOT a product of enzyme-catalyzed β-oxidation.
Tuesday, October 29, 2013
The Khan Academy and AAMC Teach the Central Dogma of Molecular Biology in Preparation for the MCAT
Here's a presentation by Tracy Kovach, a 3rd year medical student at the University of Virginia School of Medicine. Sandwalk readers will be familiar with my view of Basic Concepts: The Central Dogma of Molecular Biology and the widespread misunderstanding of Crick's original idea. It won't be a surprise to learn that a 3rd year medical student is repeating the old DNA to RNA to protein mantra.
I suppose that's excusable, especially since that's what is likely to be tested on the MCAT. I wonder if students who take my course, or similar courses that correctly teach the Central Dogma, will be at a disadvantage on the MCAT?
The video is posted on the Khan Academy website at: Central dogma of molecular biology. What I found so astonishing about the video presentation is that Tracy Kovach spends so much time explaining how to remember "transcription" and "translation" and get them in the right order. Recall that this video is for students who are about to graduate from university and apply to medical school. I expect high school students to have mastered the terms "transcription" and "translation." I'm pretty sure that students in my undergraduate class would be insulted if I showed them this video. They would be able to describe the biochemistry of transcription and translation in considerable detail.
There are people who think that the Central Dogma is misunderstood to an even greater extent than I claim. They say that the Central Dogma is widely interpreted to mean that the only role of DNA information is to make RNA which makes protein. In other words, they fear that belief in that version of the Central Dogma rules out any other role for DNA. This is the view of John Mattick. He says that the Central Dogma has been overthrown by the discovery of genes that make functional RNA but not protein.
I wonder if students actually think that this is what the Central Dogma means? Watch the first few minutes of the video and give me your opinion. Is this what she is saying?
I suppose that's excusable, especially since that's what is likely to be tested on the MCAT. I wonder if students who take my course, or similar courses that correctly teach the Central Dogma, will be at a disadvantage on the MCAT?
The video is posted on the Khan Academy website at: Central dogma of molecular biology. What I found so astonishing about the video presentation is that Tracy Kovach spends so much time explaining how to remember "transcription" and "translation" and get them in the right order. Recall that this video is for students who are about to graduate from university and apply to medical school. I expect high school students to have mastered the terms "transcription" and "translation." I'm pretty sure that students in my undergraduate class would be insulted if I showed them this video. They would be able to describe the biochemistry of transcription and translation in considerable detail.
There are people who think that the Central Dogma is misunderstood to an even greater extent than I claim. They say that the Central Dogma is widely interpreted to mean that the only role of DNA information is to make RNA which makes protein. In other words, they fear that belief in that version of the Central Dogma rules out any other role for DNA. This is the view of John Mattick. He says that the Central Dogma has been overthrown by the discovery of genes that make functional RNA but not protein.
I wonder if students actually think that this is what the Central Dogma means? Watch the first few minutes of the video and give me your opinion. Is this what she is saying?
The Khan Academy and the Association of American Medical Colleges (AAMC) Team Up to Teach Evolution and Biochemistry for the New MCAT
Theme
Better BiochemistryStudents have to write an exam called the MCAT in order to get into American Medical Schools (Canadians students also write the MCAT). The exam is created and marked by the Association of American Medical Colleges (AAMC). The format of the exam is changing in 2015 to include more biochemistry and molecular biology. This means that "pre-med" students will likely be taking more biochemistry and molecular biology courses.
Most American schools teach to the MCAT in their biochemistry and molecular biology courses because there are large numbers of wannna-be doctors in their class. The biochemistry lecturers feel that it's their duty to prep the pre-med students to pass the MCAT. This has a devastating effect on American biochemistry courses [Better Biochemistry: Teaching to the MCAT?] [Better Biochemistry: Teaching ATP Hydrolysis for the MCAT]. It is inconsistent with the American Society for Biochemistry and Molecular Biology (ASBM) goals of developing concept-driven courses that focus on fundamental principles [Fundamental Concepts in Biochemistry and Molecular Biology ].
The Khan Academy is taking advantage of the new MCAT in 2015 by posting a series of videos on basic biochemistry and evolution. The content is approved by the AAMC in order to make sure it is suitable for MCAT preparation. Here's what they say on their website [Khan Academy MCAT].
I'm going to look at the videos on evolution prepared by a second year MD/PhD student at Albert Einstein College of Medicine and videos on biochemistry prepared by a second year MD student at Harvard Medical School and a third year med student at the University of Virginia School of Medicine. Medical students are very bright and very confident of their abilities. We'll see if these students learned enough in their undergraduate courses to be able to create accurate videos that will help university graduates pass the MCAT.
Before looking at some specific examples, let me make a general comment on Khan Academy videos. I've looked at quite a few of them over the years and every single one I've seen is a "kindergarten-level" video. What I mean by that is that the level of the presentation is barely suitable for students beginning high school and in some cases they really are pitched at the level my three-year old granddaughter could understand in a year or two. They certainly aren't up to the level of any university course that I've ever taught.
These MCAT videos are no exception. But they are intended for students who are about to graduate from university. Most of these students will be getting a science degree. The mini courses are intended for students who are about to write the MCAT exam and this should represent the level of knowledge expected of medical students. As a general rule, the students who are preparing for the MCAT have achieved high grades in their biology and chemistry courses and in their biochemistry and molecular biology courses. They wouldn't be considering medical school if they weren't in the top 25% of their class.
Why are the videos pitched at such a low level of education? Is this truly representative of the quality of university education in American universities? Check them out for yourself at: Biomolecules.
Better BiochemistryStudents have to write an exam called the MCAT in order to get into American Medical Schools (Canadians students also write the MCAT). The exam is created and marked by the Association of American Medical Colleges (AAMC). The format of the exam is changing in 2015 to include more biochemistry and molecular biology. This means that "pre-med" students will likely be taking more biochemistry and molecular biology courses.
Most American schools teach to the MCAT in their biochemistry and molecular biology courses because there are large numbers of wannna-be doctors in their class. The biochemistry lecturers feel that it's their duty to prep the pre-med students to pass the MCAT. This has a devastating effect on American biochemistry courses [Better Biochemistry: Teaching to the MCAT?] [Better Biochemistry: Teaching ATP Hydrolysis for the MCAT]. It is inconsistent with the American Society for Biochemistry and Molecular Biology (ASBM) goals of developing concept-driven courses that focus on fundamental principles [Fundamental Concepts in Biochemistry and Molecular Biology ].
The Khan Academy is taking advantage of the new MCAT in 2015 by posting a series of videos on basic biochemistry and evolution. The content is approved by the AAMC in order to make sure it is suitable for MCAT preparation. Here's what they say on their website [Khan Academy MCAT].
This collection is being developed for the revised MCAT® exam that will first be administered in spring 2015. Videos will be added to the collection through fall 2014. All content in this collection has been created under the direction of the Khan Academy and has been reviewed under the direction of the Association of American Medical Colleges (AAMC). All materials are categorized according to the pre-health competencies tested by the MCAT²⁰¹⁵ exam; however, the content in this collection is not intended to prescribe a program of study for the MCAT²⁰¹⁵ exam. The content is also included in the Pre-health Collection within MedEdPORTAL’s iCollaborative sponsored by the AAMC: www.mededportal.org/pre-health *MCAT® is a program of the AAMC and related trademarks owned by the Association include Medical College Admission Test, MCAT, and MCAT²⁰¹⁵. For more information about the MCAT exam visit : www.aamc.org/mcat2015.So, how did the Khan Academy prepare the videos? They set up an MCAT Video Competition and picked the best ones. You can read about the winners at MCAT Video Competition Winners. It's an eclectic mix of people but 11 out of 15 winners are medical school students or graduate students. Keep in mind that teaching introductory subjects like evolution and biochemistry is hard and Teachers Have to Know Their Subject.
I'm going to look at the videos on evolution prepared by a second year MD/PhD student at Albert Einstein College of Medicine and videos on biochemistry prepared by a second year MD student at Harvard Medical School and a third year med student at the University of Virginia School of Medicine. Medical students are very bright and very confident of their abilities. We'll see if these students learned enough in their undergraduate courses to be able to create accurate videos that will help university graduates pass the MCAT.
Before looking at some specific examples, let me make a general comment on Khan Academy videos. I've looked at quite a few of them over the years and every single one I've seen is a "kindergarten-level" video. What I mean by that is that the level of the presentation is barely suitable for students beginning high school and in some cases they really are pitched at the level my three-year old granddaughter could understand in a year or two. They certainly aren't up to the level of any university course that I've ever taught.
These MCAT videos are no exception. But they are intended for students who are about to graduate from university. Most of these students will be getting a science degree. The mini courses are intended for students who are about to write the MCAT exam and this should represent the level of knowledge expected of medical students. As a general rule, the students who are preparing for the MCAT have achieved high grades in their biology and chemistry courses and in their biochemistry and molecular biology courses. They wouldn't be considering medical school if they weren't in the top 25% of their class.
Why are the videos pitched at such a low level of education? Is this truly representative of the quality of university education in American universities? Check them out for yourself at: Biomolecules.
Monday, October 28, 2013
Michael Egnor Keeps Digging
When you find yourself in a hole, stop digging.
Will RogersI favor teaching biochemistry from an evolutionary perspective and I was pleased to see that ASBMB considers evolution to be one of the fundamental concepts in biochemistry and molecular biology [ASBMB Core Concepts in Biochemistry and Molecular Biology: Evolution]. (ASBMB screws up their description of evolution but at least their heart's in the right place.)
Unless they understand evolution, students can't really understand why some parts of a protein are the same in all species and other parts are quite variable. They certainly can't understand why you can construct a phylogenetic tree from sequences and why this tree closely resembles those trees made from comparing anatomy/embryology. They won't know why those molecular trees are consistent with a fossil record unless they understand evolution.
Read more »
Will RogersI favor teaching biochemistry from an evolutionary perspective and I was pleased to see that ASBMB considers evolution to be one of the fundamental concepts in biochemistry and molecular biology [ASBMB Core Concepts in Biochemistry and Molecular Biology: Evolution]. (ASBMB screws up their description of evolution but at least their heart's in the right place.)
Unless they understand evolution, students can't really understand why some parts of a protein are the same in all species and other parts are quite variable. They certainly can't understand why you can construct a phylogenetic tree from sequences and why this tree closely resembles those trees made from comparing anatomy/embryology. They won't know why those molecular trees are consistent with a fossil record unless they understand evolution.
Read more »
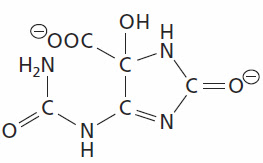
Monday's Molecule #221
Last week's molecule was 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline (OHCU). It is an intermediate in the degradation pathway from uric acid (or urate) to carbon dioxide and ammonia. Uric acid is the main breakdown product in purine catabolism. Humans have lost activity of all of the enzymes of this pathway so they excrete urate. Most other species excrete ammonia, although in other animals some of the terminal enzymes have been lost.
Some textbooks do not show the uric acid degradation pathway since it doesn't occur in humans and those textbooks aren't interested in an evolutionary approach to biochemistry (e.g. Berg, Tymoczko, and Stryer). The other majors textbooks (Voet & Voet, Garrett & Grisham, Nelson & Cox [Lehinger]) all show uric acid converted directly to allantoin via urate oxidase. This reaction was shown to be incorrect about 15 years ago. The actual pathway from uric acid to allantoin involves two intermediates; 5-hydroxyisourate and OHCU.
 Jean-Marc was kind enough to send me a menu [PDF]. There are about 30 different fondues to choose from. If you would like to join us you can leave a comment on last week's post.
Jean-Marc was kind enough to send me a menu [PDF]. There are about 30 different fondues to choose from. If you would like to join us you can leave a comment on last week's post.
This week's molecule is related to a discussion we are having on the How Do the IDiots Explain the Origin of Life? post. Can you identify this molecule? You have to be very specific.
Email your answer to me at: Monday's Molecule #221. I'll hold off posting your answers for at least 24 hours. The first one with the correct answer wins. I will only post the names of people with mostly correct answers to avoid embarrassment. The winner will be treated to a free lunch.
There could be two winners. If the first correct answer isn't from an undergraduate student then I'll select a second winner from those undergraduates who post the correct answer. You will need to identify yourself as an undergraduate in order to win. (Put "undergraduate" at the bottom of your email message.)
Read more »
Some textbooks do not show the uric acid degradation pathway since it doesn't occur in humans and those textbooks aren't interested in an evolutionary approach to biochemistry (e.g. Berg, Tymoczko, and Stryer). The other majors textbooks (Voet & Voet, Garrett & Grisham, Nelson & Cox [Lehinger]) all show uric acid converted directly to allantoin via urate oxidase. This reaction was shown to be incorrect about 15 years ago. The actual pathway from uric acid to allantoin involves two intermediates; 5-hydroxyisourate and OHCU.
Image Credit: Moran, L.A., Horton, H.R., Scrimgeour, K.G., and Perry, M.D. (2012) Principles of Biochemistry 5th ed., Pearson Education Inc. page 568 [Pearson: Principles of Biochemistry 5/E] © 2012 Pearson Education Inc.The winner, for the second week in a row, is Jean-Marc Neuhaus. [Monday's Molecule #220]. Jean-Marc lives in Switzerland so I've made arrangements to fly over there to visit him and treat him to two fondues at the Pinte de Pierre-à-Bot in Neuchatel.
 Jean-Marc was kind enough to send me a menu [PDF]. There are about 30 different fondues to choose from. If you would like to join us you can leave a comment on last week's post.
Jean-Marc was kind enough to send me a menu [PDF]. There are about 30 different fondues to choose from. If you would like to join us you can leave a comment on last week's post. This week's molecule is related to a discussion we are having on the How Do the IDiots Explain the Origin of Life? post. Can you identify this molecule? You have to be very specific.
Email your answer to me at: Monday's Molecule #221. I'll hold off posting your answers for at least 24 hours. The first one with the correct answer wins. I will only post the names of people with mostly correct answers to avoid embarrassment. The winner will be treated to a free lunch.
There could be two winners. If the first correct answer isn't from an undergraduate student then I'll select a second winner from those undergraduates who post the correct answer. You will need to identify yourself as an undergraduate in order to win. (Put "undergraduate" at the bottom of your email message.)
Read more »
Saturday, October 26, 2013
How to Turn a Simple Paper into a Scientific Breakthrough: Mention Junk DNA
Attanasio et al. (2013) published a paper in Science where they identified several thousand possible enhancers that were active in the facial area of developing mouse embryos. About 200 of them appear to be controlling genes that determine the size and shape of the face. (Recall that there are about 20,000 protein-encoding genes in mammals.)
Lynn Yarris of Lawrence Berkeley National Laboratory in California (USA) wrote up the press release [What is it About Your Face?]. It's a really good press release that fairly represents the published work and explains some of the significance. There's no mention of junk DNA in the press release or the published paper.
This is what it looks like when science correspondent Alok Jha published it in The Guardian.
Scientists have known for decades that a lot of noncoding DNA is functional. The idea that all noncoding DNA (98%) is junk is false. No knowledgeable scientist ever made such a claim. It is a myth perpetuated, in part, by ignorant science writers; albeit, aided and abetted by ignorant scientists. Scientists have known for fifty (50!!) years that gene expression is controlled by regulatory sequences in noncoding DNA. Scientists have known for at least that length of time that during embryogenesis different genes are turned on and off and that this is due, in part, to binding of transcription factors to those regulatory sequences (enhancers). Scientists have known for one hundred years that the morphological features of mammals, including humans, are controlled by genes.
Move along folks. There's nothing to see here.
Lynn Yarris of Lawrence Berkeley National Laboratory in California (USA) wrote up the press release [What is it About Your Face?]. It's a really good press release that fairly represents the published work and explains some of the significance. There's no mention of junk DNA in the press release or the published paper.
This is what it looks like when science correspondent Alok Jha published it in The Guardian.
Faces are sculpted by 'junk DNA'It's pretty clear that science correspondent Alok Jha doesn't understand what he's writing and it's about time we started publicizing the names of those science writers who mislead the public about science. The consensus among knowledgeable scientists is that at least 80-90% of our genome is junk. It's time for science writers to admit that the science favors junk.
Though everybody's face is unique, the actual differences are relatively subtle. What distinguishes us is the exact size and position of things like the nose, forehead or lips. Scientists know that our DNA contains instructions on how to build our faces, but until now they have not known exactly how it accomplishes this.
Visel's team was particularly interested in the portion of the genome that does not encode for proteins – until recently nicknamed "junk" DNA – but which comprises around 98% of our genomes. In experiments using embryonic tissue from mice, where the structures that make up the face are in active development, Visel's team identified more than 4,300 regions of the genome that regulate the behaviour of the specific genes that code for facial features.
Scientists have known for decades that a lot of noncoding DNA is functional. The idea that all noncoding DNA (98%) is junk is false. No knowledgeable scientist ever made such a claim. It is a myth perpetuated, in part, by ignorant science writers; albeit, aided and abetted by ignorant scientists. Scientists have known for fifty (50!!) years that gene expression is controlled by regulatory sequences in noncoding DNA. Scientists have known for at least that length of time that during embryogenesis different genes are turned on and off and that this is due, in part, to binding of transcription factors to those regulatory sequences (enhancers). Scientists have known for one hundred years that the morphological features of mammals, including humans, are controlled by genes.
Move along folks. There's nothing to see here.
Attanasio, C. et al. (2013) Fine Tuning of Craniofacial Morphology by Distant-Acting Enhancers. Science 342: Oct. 25, 2013 [doi: 10.1126/science.1241006]
Thursday, October 24, 2013
ASBMB Core Concepts in Biochemistry and Molecular Biology: Matter and Energy Transformation
Theme
Better BiochemistryTansey et al. (2013) have described the five core concepts in biochemistry and molecular biology. These are the fundamental concepts that all biochemistry instructors must teach and all biochemistry students must understand.
The five core concept categories are:
Let's see how they do with the second core concept.
First, I would have mentioned that organisms can capture energy from simple inorganic compounds such as H2 or those containing Fe2+. These are energy sources for many chemoautrophic bacteria. If you are teaching biochemistry from an evolutionary perspective, it's important that students understand how these organisms capture energy. That's the process that is most like the mechanism found in the earliest living cells.1
Second, I would have put more emphasis on using captured energy in biosynthesis pathways. The paragraph mentions that energy can be used to generate new chemical bonds but that doesn't convey the importance of the process. Think about bacterial cells growing and dividing in the ocean or plants growing from a single seed. Most of the energy goes into making proteins, nucleic acids, lipids, and carbohydrates.
Third, I would drop the reference to cells being in "a state of nonequilibrium with their environment." That conceptt is covered under "homeostasis."
Not only that, what does it mean to say that a reaction is "energetically unfavorable"? Usually this refers to the standard Gibbs free energy (ΔG°′) but one of the most important concepts in biochemistry is the difference between the standard Gibbs free energy change and the actual Gibbs free energy change (ΔG) inside the cell. In most cases ΔG = 0.
It's true that there are potential "endergonic" reactions occurring inside cells. Think about ATP hydrolysis, for example. The concentration of ATP is maintained at a high level relative to ADP and Pi so the Gibbs free energy change in the direction of hydrolysis is actually more negative that even the standard Gibbs free energy change. What this means is the the reverse reaction is extremely "endergonic."
However, it is simply not true that there are steps in metabolic pathways that are "endergonic" as the authors state. That statement reflects a profound misunderstanding of a fundamental concept in biochemistry. There will not be any flux in the "forward" direction of a metabolic pathway as long as even one reaction is "endergonic." All reactions have to be near-equilibrium reactions or reactions with a negative ΔG that's maintained because the enzyme activity is regulated to prevent the reaction from reaching equilibrium.
The important concept is "flux" or flow of metabolites in one direction along a metabolic pathway. There are many pathways where flux can occur in either direction as in the central part of the gluconeogenesis/glycolysis pathway or the citric acid cycle. Students need to understand what controls flux in one direction or another. They should know that, like water, metabolic flux cannot flow uphill.
The point is that the enzyme (glutamine synthetase) catalzyes a completely different reaction—a phosphoryl group transfer reaction—with a negative standard Gibbs free energy change of ΔG°′ = −18 kJ mol-1.
[see Moran et al. (2011): Introduction to Metabolism]
If there were an enzyme that catalyzed the first reaction involving only glutamate and ammonia then this reaction could easily occur inside the cell in spite of the positive ΔG°′. It would be a near-equilibrium reaction with steady-state equilibrium concentrations of glutamate that were very much higher than the concentration of glutamine.
It's likely that the concentration of glutamine would then be too low to support all the reactions that require it. That's why the reaction involving ATP is more useful. It means that the steady-state concentration of glutamine can be maintained a much higher concentration. This requires regulation of glutamine synthetase in order to prevent the reaction from reaching equilibrium.
It seems to me that the authors (Tansey et al.) have not thought about the fundamental core concepts. They are promoting widespread misconceptions about thermodynamics and metabolism and they are missing some important concepts. I've already mentioned flux. The other missing concept is oxidation-reduction reactions (electron transfer) and the importance of reduction potentials. NADH, NADPH, and QH2 are important energy currencies inside the cell—just as important as ATP.
There's something seriously wrong with biochemistry teaching if ASBMB educators can't even correctly explain foundational concepts like "evolution" and "matter and energy transformation."
Better BiochemistryTansey et al. (2013) have described the five core concepts in biochemistry and molecular biology. These are the fundamental concepts that all biochemistry instructors must teach and all biochemistry students must understand.
The five core concept categories are:
- Evolution
- matter and energy transformation
- homeostasis
- biological information
- macromolecular structure and function
Let's see how they do with the second core concept.
Matter and Energy TransformationI think we can all agree that a basic understanding of thermodynamics is an important core concept. However, I would have worded this paragraph somewhat differently.
The Many Forms of Energy Involved in Biological Processes
The energetics of a biological system or process—be it an ecosystem, an organism, a cell, a biochemical reaction—conforms to and is understood in terms of the fundamental laws of thermodynamics. Biological systems capture and process energy from the environment in many forms including that emanating directly from the sun (photons through photosynthesis), heat from the environment (kinetic energy), and energy rich compounds produced by geothermal processes (e.g. sulfur compounds) or other organisms (e.g. carbohydrates). Energy from all sources is chemically converted into useful chemical and physical work in a controlled and regulated fashion. The potential
energy stored in chemical bonds can used to generate motion, light, heat, and electrochemical gradients; likewise, electrochemical gradients can be used to generate motion and new chemical bonds. The input of energy from the environment allows living systems to exist in a state of nonequilibrium with their environment. The discussion of energy and matter conversions in biological systems makes use of the physical concept of changes in Gibbs free energy, or ΔG.
First, I would have mentioned that organisms can capture energy from simple inorganic compounds such as H2 or those containing Fe2+. These are energy sources for many chemoautrophic bacteria. If you are teaching biochemistry from an evolutionary perspective, it's important that students understand how these organisms capture energy. That's the process that is most like the mechanism found in the earliest living cells.1
Second, I would have put more emphasis on using captured energy in biosynthesis pathways. The paragraph mentions that energy can be used to generate new chemical bonds but that doesn't convey the importance of the process. Think about bacterial cells growing and dividing in the ocean or plants growing from a single seed. Most of the energy goes into making proteins, nucleic acids, lipids, and carbohydrates.
Third, I would drop the reference to cells being in "a state of nonequilibrium with their environment." That conceptt is covered under "homeostasis."
CatalysisThis is pretty good. I would only add that there are some fundamental concepts of enzyme mechanisms that need to be covered. The idea of a transition state is important. I put a lot of emphasis on oxidation-reduction reactions as a core concept in biochemistry.
Biologically relevant energy and matter interconversions do not occur rapidly enough (often by many orders of magnitude) to support life. In living systems, biological catalysts called enzymes facilitate these reactions. Enzymes are macromolecules, usually proteins or RNA molecules with a catalytic function. Enzymes do not alter reaction equilibria; instead, they lower the activation barrier of a particular reaction so that reactions proceed much more rapidly. The presence of powerful enzymatic catalysts is one of the key conditions for life itself.
Description of the rates of enzymatic reactions represents the subdiscipline enzyme kinetics. Key concepts of kinetics, including the definitions of the terms vo, Vmax, Km, and kcat, constitute a common language for biochemists and molecular biologists in discussing the properties of enzymes.
Students should be able to apply their knowledge of basic chemical thermodynamics to biologically catalyzed systems, quantitatively model how these reactions occur, and calculate kinetic parameters from experimental data.
Coupling Exergonic and Endergonic ProcessesI have a problem with this section. I don't think that the concepts of "exergonic" and "endergonic" processes are very important in biochemistry and I don't use them in my textbook. They're not found in many other textbooks, either. Also, the idea of "coupled" reactions is very poorly taught in biochemistry courses. It's almost never true that enzymes simply link up two independent reactions, one of which is "favorable" and the other "unfavorable." What usually happens is that a completely new reaction is catalyzed. For example, ATP is not hydrolyzed but, instead, a group transfer reaction is created. This important concept is covered in the next section but the authors do not appear to have grasped its significance.
Biochemical systems couple energetically unfavorable reactions with energetically favorable reactions to allow for a wider variety of reactions to proceed.
Students should be able to discuss the concept of Gibbs free energy, and how to apply it to chemical transformations, be able to identify which steps of metabolic pathways are exergonic and which are endergonic and relate the energetics of the reactions to each other.
Not only that, what does it mean to say that a reaction is "energetically unfavorable"? Usually this refers to the standard Gibbs free energy (ΔG°′) but one of the most important concepts in biochemistry is the difference between the standard Gibbs free energy change and the actual Gibbs free energy change (ΔG) inside the cell. In most cases ΔG = 0.
It's true that there are potential "endergonic" reactions occurring inside cells. Think about ATP hydrolysis, for example. The concentration of ATP is maintained at a high level relative to ADP and Pi so the Gibbs free energy change in the direction of hydrolysis is actually more negative that even the standard Gibbs free energy change. What this means is the the reverse reaction is extremely "endergonic."
However, it is simply not true that there are steps in metabolic pathways that are "endergonic" as the authors state. That statement reflects a profound misunderstanding of a fundamental concept in biochemistry. There will not be any flux in the "forward" direction of a metabolic pathway as long as even one reaction is "endergonic." All reactions have to be near-equilibrium reactions or reactions with a negative ΔG that's maintained because the enzyme activity is regulated to prevent the reaction from reaching equilibrium.
The important concept is "flux" or flow of metabolites in one direction along a metabolic pathway. There are many pathways where flux can occur in either direction as in the central part of the gluconeogenesis/glycolysis pathway or the citric acid cycle. Students need to understand what controls flux in one direction or another. They should know that, like water, metabolic flux cannot flow uphill.
The Nature of Biological EnergyThe essence of these statements is correct but it is not explained very well. The important concept is not that you "couple" a "favorable" reaction like ATP hydrolysis to an "unfavorable" reaction like synthesis of glutamine from glutamate and ammonia (ΔG°′ = +14 kJ mol-1).
In biological systems, chemical energy is stored in molecules with high group transfer potential or strongly negative free energy of hydrolysis or decomposition. These molecules, particularly ATP, provide the free energy to drive otherwise unfavorable biochemical reactions or processes in tightly coupled and highly controlled fashion. Most frequently, the free energy needed for a process or metabolic pathway is provided by group transfer rather than by hydrolysis. In this way, efficient energy transfer is optimized, while inefficient energy transfer to the environment (in the form of heat for example) is minimized.
Students should be able to show how reactions that proceed with large negative changes in free energy can be used to render other biochemical processes more favorable.
The point is that the enzyme (glutamine synthetase) catalzyes a completely different reaction—a phosphoryl group transfer reaction—with a negative standard Gibbs free energy change of ΔG°′ = −18 kJ mol-1.
[see Moran et al. (2011): Introduction to Metabolism]
If there were an enzyme that catalyzed the first reaction involving only glutamate and ammonia then this reaction could easily occur inside the cell in spite of the positive ΔG°′. It would be a near-equilibrium reaction with steady-state equilibrium concentrations of glutamate that were very much higher than the concentration of glutamine.
It's likely that the concentration of glutamine would then be too low to support all the reactions that require it. That's why the reaction involving ATP is more useful. It means that the steady-state concentration of glutamine can be maintained a much higher concentration. This requires regulation of glutamine synthetase in order to prevent the reaction from reaching equilibrium.
It seems to me that the authors (Tansey et al.) have not thought about the fundamental core concepts. They are promoting widespread misconceptions about thermodynamics and metabolism and they are missing some important concepts. I've already mentioned flux. The other missing concept is oxidation-reduction reactions (electron transfer) and the importance of reduction potentials. NADH, NADPH, and QH2 are important energy currencies inside the cell—just as important as ATP.
There's something seriously wrong with biochemistry teaching if ASBMB educators can't even correctly explain foundational concepts like "evolution" and "matter and energy transformation."
1. I believe that all introductory biochemistry students should be able to explain where chemoautrophs get their energy. If they can't do it, they haven't been taught the fundamental concepts.
Tansey, J.T., Baird, T., Cox, M.M., Fox, K.M., Knight, J., Sears, D. and Bell, E. (2013) Foundational concepts and underlying theories for majors in “biochemistry and molecular biology”. Biochem. Mol. Biol. Educ., 41:289–296. [doi: 10.1002/bmb.20727]
Monday, October 21, 2013
Evolution Is Irrelevant to Michael Egnor
The title of this post suggest a story that's about as interesting as the proverbial "Dog Bites Man" story [see Man Bites Dog]. Nevertheless, from time to time it is amusing to see how the creationist mind works.
Michael Egnor is upset about the fact that the American Society for Biochemistry and Moleclar Biology (ASBMB) picked "evolution" as an important concept that should be covered in a biochemistry or molecular biology course. He doesn't like my post: ASBMB Core Concepts in Biochemistry and Molecular Biology: Evolution. He decided that he better convince his fellow creationists than biochemists don't know what they are talking about [Is Darwinian Evolution "Indispensable" to Biology?].
Here are some excerpts for your amusement.
Read more »
Michael Egnor is upset about the fact that the American Society for Biochemistry and Moleclar Biology (ASBMB) picked "evolution" as an important concept that should be covered in a biochemistry or molecular biology course. He doesn't like my post: ASBMB Core Concepts in Biochemistry and Molecular Biology: Evolution. He decided that he better convince his fellow creationists than biochemists don't know what they are talking about [Is Darwinian Evolution "Indispensable" to Biology?].
Here are some excerpts for your amusement.
Read more »
Monday's Molecule #220
Last week's molecule was citrate synthase, one of many enzymes that show considerable amounts of structural change during binding. It looks like the "induced fit" mechanism is a general feature of substrate binding and not something that is limited to just a few examples. That part of the question was easy but the second part was hard. Jean-Marc Neuhaus is this week's winner because he has a copy of my book and was able to look up the explanation. The important point to keep in mind when you are thinking about the thermodynamics of biochemical reactions is that most reactions are near-equilibrium reactions where ΔG = 0. In the case of the citrate synthase reaction, ΔG°′ = -31.5 kJ mol-1, in the direction of citrate formation. What this means is that the equilibrium concentrations of the products are very much higher than the concentrations of the substrates. These concentrations would be closer to being equal if the reaction was coupled to substrate level phosphorylation (e.g. ATP formation). This would be a problem since the concentration of oxaloacetate (substrate) inside the cell is very low. (Because the standard free energy change of the malate dehydrogenase reaction is ΔG°′ = +30 kJ mol-1) [Monday's Molecule #219]. Jean-Marc lives in Switzerland so I've made arrangements to fly over there to visit him and treat him to fondue at the Pinte de Pierre-à-Bot in Neuchatel. 
Today's molecule is one of those molecules that students should never be asked to memorize. It's an intermediate in a very important pathway. Identify the molecule and the pathway. You have to give me the full name and the common abbreviation. Email your answer to me at: Monday's Molecule #220. I'll hold off posting your answers for at least 24 hours. The first one with the correct answer wins. I will only post the names of people with mostly correct answers to avoid embarrassment. The winner will be treated to a free lunch.
There could be two winners. If the first correct answer isn't from an undergraduate student then I'll select a second winner from those undergraduates who post the correct answer. You will need to identify yourself as an undergraduate in order to win. (Put "undergraduate" at the bottom of your email message.)
Read more »

Today's molecule is one of those molecules that students should never be asked to memorize. It's an intermediate in a very important pathway. Identify the molecule and the pathway. You have to give me the full name and the common abbreviation. Email your answer to me at: Monday's Molecule #220. I'll hold off posting your answers for at least 24 hours. The first one with the correct answer wins. I will only post the names of people with mostly correct answers to avoid embarrassment. The winner will be treated to a free lunch.
There could be two winners. If the first correct answer isn't from an undergraduate student then I'll select a second winner from those undergraduates who post the correct answer. You will need to identify yourself as an undergraduate in order to win. (Put "undergraduate" at the bottom of your email message.)
Read more »
Subscribe to:
Posts (Atom)